Safety of the antigen of the Luebeck anti-Corona vaccination
The Lübeck corona antigen is a genetically engineered copy of the receptor-binding domain of the corona virus (Fig. 1). Already since the 26th of March 2020, it has been successfully used in Lübeck for vaccination against Covid-19. Some public authorities have criticized the fact that GMP-compliant production cannot be demonstrated for the corona protein used in the Lübeck vaccination. In terms of urgency, this is the wrong approach, as it takes at least two years to establish GMP-compliant production. But the world needs effective vaccines today, not in two years, and with a non-hazardous product. This is not the time for a large-scale global experiment with a new vaccination principle that has not yet been sufficiently tested. Instead, methods must be used that have been tried and tested a hundred million times: An antigen is applied that has been synthesized and that the body does not have to produce itself first, whereby it would itself become the target of the emerging immunity.
Under my supervision, the quality of the Lübeck vaccine is closely controlled. It has the required quality, but for formal reasons it does not yet have a corresponding official certificate. Even without such a certificate, the safety of the Lübeck vaccine has long been proven in 50,000 vaccinations. It is effective and does not make anyone ill, and according to the German Basic Law, such proof should be dispensable as long as the vaccine is produced by the doctors themselves and administered individually at the request of each patient after appropriate information. However, it may not be marketed, i.e. sold to doctors who do not produce it themselves., According to surveys, 10 million people in Germany would immediately be voluntarily vaccinated with the Lübeck method, and no sanctions would be required to achieve complete herd immunity against Corona virus.
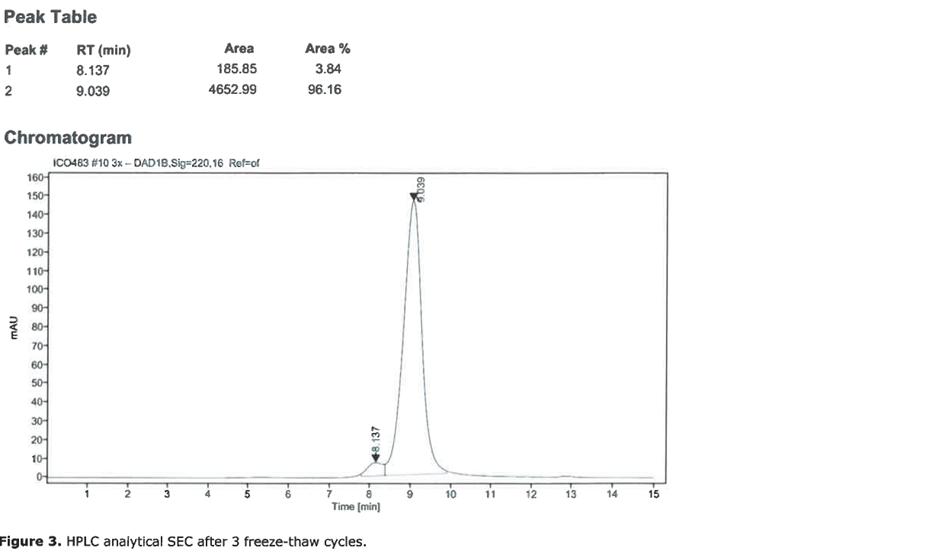
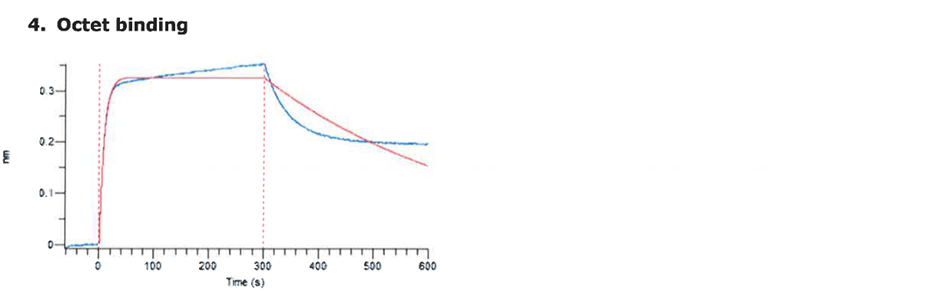
For the production of the recombinant SARS-CoV2 spike RBD2 protein (Fig. 1), the CHOEBNALT-85 system is used (Table 1). Cell culture of this CHO-based cell line (chinese hamster ovary) is performed in serum-free culture medium. The CHO cell line was stated by the manufacturer to be free of retroviruses. The protein is isolated by metal affinity chromatography followed by size exclusion chromatography and passed through a 0.2-µm sterile filter. Protein identity was confirmed by mass spectroscopy (Table 2, Figure 2). It shows high purity (>95%, based on SDS-PAGE, Figure 3), high monodispersity (analytical size exclusion chromatography, Figure 4 top) and high stability (analytical size exclusion chromatography after 3 freeze-thaw cycles, Figure 4 bottom). The functionality of the SARS-CoV2 spike RBD2 was validated by determining the binding affinity to the ACE2 receptor using Octet measurements (Figure 5).
In addition, the neutralizing effect of the antigen on anti-Covid-19 positive patient sera is also measured (Figure 6).
Due to the preparation methods used, particles larger than the respective RBD protein (hydrodynamic diameter: approx. 2.5nm) are separated. Thus, a possible contamination of retroviruses or parvoviruses (hydrodynamic diameter: approx. 100nm and 25nm, respectively) is excluded in the final product.
The measured endotoxin concentration according to the Limulus amebocyte lysate test (LAL test) of the SARS-CoV2 spike RBD2 is below 0.1EU/mg (see Figure 1, manufacturer’s specifications).
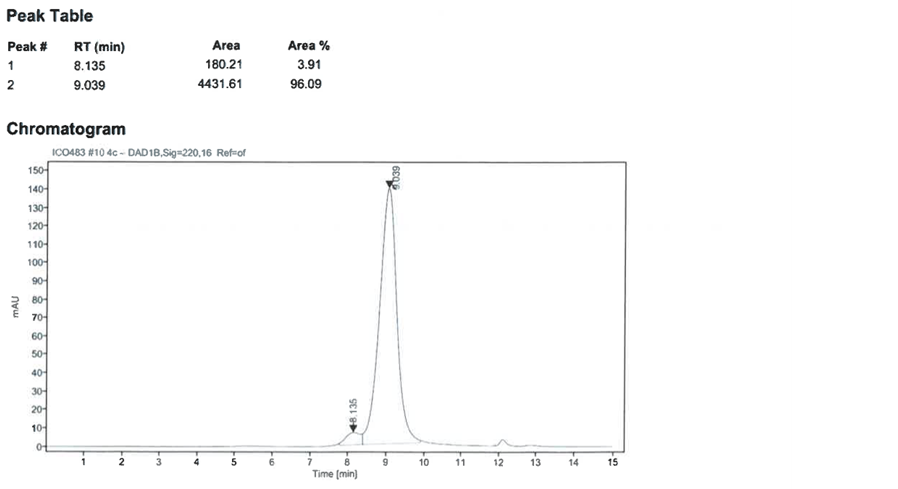
No microbiological growth was detected in the sterility test (bacteriological) of the protein (see figure 7).
Figure 1: Structure of the recombinant SARS-CoV2 spike RBD2 protein
Table 1: Manufacturer’s specifications for SARS-CoV-2 S1 RBD2
In the protein analysis, the score was calculated by the software Mascot with -10*log(P), where P is the probability that the hit found is a random event. A score above the cut-off is considered significant (p<0.05). If the target antigen was found during the evaluation, it is indicated in the table. In addition, all unrelated proteins above the cut-off are indicated, taking into account origin (e.g. organism, production) and size. If there were no hits above the cut-off, the band was labelled “not identified”. Database: combined sub-database of the NCBInr database (mammalia + viruses combined).
Table 2: Mass spectrometric protein analysis of the SARS-CoV-2 S1 RBD2
After reduction and alkylation, SARS-CoV-2 S1 RBD2 was denatured using lithium dodecyl sulfate followed by electrophoretic separation in Bis-Tris gel (MES buffer system). Staining was performed using Blue Silver. The labelled bands (41-44) were cut out of the gel, proteolysed in the gel using trypsin and then analysed by MALDI-TOF.
The mass spectrometric analysis of the tryptic peptides was performed on an Autoflex III (MALDI-TOF, Bruker Daltonik) using the positive reflector mode. A peptide mass fingerprint (PMF) spectrum was acquired from the peptide mixture of each gel band. Automated MS/MS analyses were performed on a maximum of four peptides from each PMF spectrum. The PMF and MS/MS data were aligned with a house-created sub-database of NCBInr (mammalia + viruses combined) using Mascot software (Matrix Science).
Figure 2: Identity determination of the SARS-CoV2 spike RBD.2
Figure 3: SDS-PAGE of the SARS-CoV-2 S1 RBD2
Figure 4: Analytical size exclusion chromatography of SARS-CoV-2 S1 RBD2. Top: initial sample, bottom: after three freeze-thaw cycles.
Figure 5: Determination of the binding affinity of SARS-CoV-2 S1 RBD2 to the ACE2 receptor using Octet measurements
The addition of RBD-delta antigen to an antibody-positive serum causes the neutralization of RBD-specific antibodies and thus leads to a decrease in signal in the anti-SARS-CoV-2 QuantiVac ELISA (IgG) (Euroimmun). Neutralization with wild-type antigen leads to analogous results.
Figure 6: Neutralization
Figure 7: Sterility testing of the SARS-CoV-2 RBD2